

Current research includes a focus on a) the development and optimization of diagnostics and enhanced sample preparation strategies for rapid detection of foodborne pathogens, b) ecology and control of foodborne pathogens, with a special emphasis on understanding and controlling antimicrobial resistance in the food supply and the environment, including wildlife, and c) development and validation of microbial source tracking approaches.

The purpose of this project is to conduct testing for the presence of SARS-CoV-2 in wastewater at the University of Wyoming in collaboration with the proposed wastewater testing efforts at the Wyoming Public Health Lab (WPHL). This will be accomplished through routine sampling of wastewater influent discharging into water treatment facilities serving several Wyoming Communities. These communities include: Cowley, Deaver, Hudson, Laramie, Pinedale, and Powell. Testing of wastewater for SARS-CoV-2 has proven to be a reliable indicator of the presence of the SARS-CoV-2 virus in communities. Wastewater testing can be more economical than clinical testing over time and can offer a snapshot of the SARS-CoV-2 burden at a community level that may not have adequate jurisdictional testing. Data generated from this project may also supplement jurisdiction that are understating testing and will provide further data for consideration of the true burden of SARS-CoV-2 within a defined region. In addition to evaluating SARS-CoV-2 in the samples through RNA extraction, DNA will also be collected. These samples will undergo sequencing, testing of the various SAR-CoV-2 variants, detection of antimicrobial resistance, and disease detection. Testing began in January of 2021 and will commence December 31st, 2021.

The World Health Organization global action plan on antimicrobial resistance (AMR) has set forth the need for integrated antibiotic resistance (AR) surveillance as one of its main objectives. Urban wastewater treatment plants (UWTPs) are recognized as receptors and sources of environmental AMR. Our goal is to understand antimicrobial resistance trends at the population level. The development of regular locally implemented surveillance protocols will support comparative analyses to determine regional and sporadic events involving emergence of priority AMR. We utilize a qPCR array with primer sets targeting antibiotic resistance genes and mobile genetic elements (MGEs). Targeted sequences comprise those involved in gene transfer and recombination, integrase, transposase, insertion sequences, plasmid replicon types, and antibiotic resistance and detection is performed by digital PCR.

Recent research at Montana State University, United States Sheep Experiment Station, and University of Wyoming has estimated the incidence of subclinical mastitis to be between 11% and 74% of ewes, much greater than previously thought. These groups reported a decrease in lamb performance of up to 35 lb per litter when reared by ewes with subclinical mastitis, which represents economic losses up to $106 per ewe. However, others have found insignificant or inconsistent findings of subclinical mastitis on lamb performance. Therefore, there is still a lack of knowledge on the extent of negative impacts associated with subclinical mastitis. Ongoing research at UW is demonstrating that ewes have a community of microbes harbored in the mammary gland throughout lactation, suggesting subclinical mastitis to be a complex disease. This past research has been partially funded by NSIIC funds and has provided valuable information, and the proposed research project will provide us the opportunity to answer the important remaining questions in this research area. Using modern and traditional methodologies to investigate the milk microbial community and milk yield in non-dairy ewes at multiple points through lactation will identify the extent and duration of the negative impacts on lamb performance, including growth and survival. The objectives of this project are to: 1) quantify the milk yield and quality reduction in ewes affected by subclinical mastitis; 2) quantify the decrease in lamb performance in those raised by ewes with subclinical mastitis; 3) characterize colonization of the lamb’s rumen microbiome to predict future lamb performance; 4) asses the efficacy and impact of antibiotics administered during early lactation to reduce mastitis; and 5) quantify effects of body weight and condition loss on somatic cell count and lamb performance.
Antimicrobial resistance (AMR) in bacteria has increasingly become a domestic and global threat to agricultural and human health. Because of their ability to transfer genetic material, once benign bacterial strains can become problematic if they acquire characteristics conferring antimicrobial resistance. In the U.S., this threat has been recognized as a national problem and a major public health concern, especially with respect to livestock production, and was recently acted upon through development of overarching strategic programs by the federal government. An essential component of effective AMR-mitigating programs is enhanced surveillance. The need for surveillance is clearly indicated in the U.S. National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) issued by the White House. Furthermore, the National Antimicrobial Resistance Monitoring System (NARMS) which is a joint program among the FDA, USDA and CDC (http://www.cdc.gov/narms/) has played a critical role in the surveillance of AMR in the animal production, the food chain, and human clinical settings in the United States. We utilize standard methods established by the retail meat arm of the NARMS to monitor foodborne bacteria in retail meats. Further, we focus on understanding links of antimicrobial resistance to wildlife, food production animals, and associated environment. By combining the data outputs of these methods, the acquired information will provide needed insight into the magnitude and causation of the AMR problem, and help determine what actions should be taken to mitigate this risk.
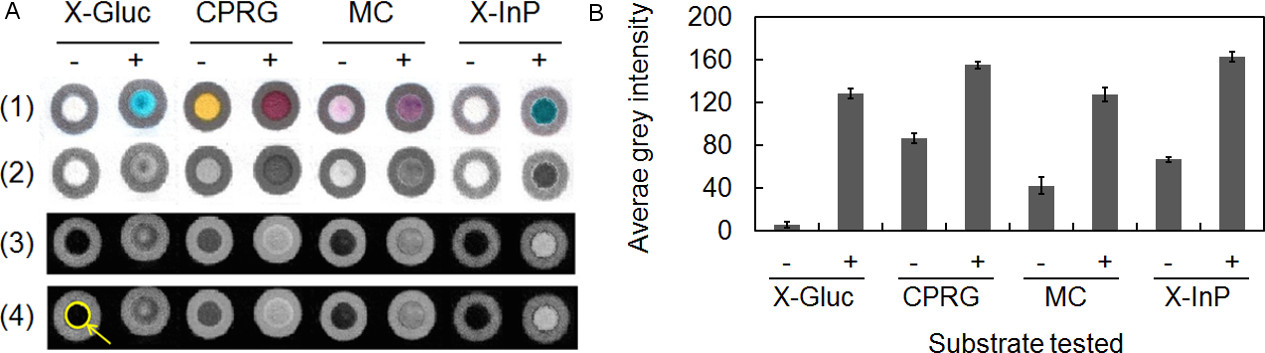
Inexpensive sensors which use paper as a support material are attractive because paper substrates are easy to use, inexpensive, and can be widely deployed. These devices, termed paper-based analytical devices (µPADs), serve as useful platforms for conducting multiplexed assays using microliter sample volumes. Because paper has a high surface area, visualization of end points is more sensitive than planar devices and increases the potential for use in minimally instrumented analysis scenarios. Through industry and academic collaborations we are developing spot array design where colorimetric assays are carried out in the “wells”, utilizing the reaction between species-indicative enzymes and chromogenic substrates.
Wyoming’s rivers and streams are host to numerous recreational activities for residents and visitors to the state, but waterborne pathogens pose a public health threat to users who come into contact with contaminated water. Pathogens levels are often indicated by certain levels of Escherichia coli and other fecal indicator bacteria (FIB); however, effective management decisions to reduce the likelihood of pathogen presence require knowledge of contaminant sources. We are advancing molecular microbial source tracking (MST) approaches involving community course tracking to accurately identify sources of contamination used in water management decisions.

Bledar Bisha is an Associate Professor in Food Microbiology and Department Head of Animal Science. His interest lie in conducting applied and basic research in food safety microbiology benefiting general consumers, public health, and the food industry. Current research includes a focus on a) the development and optimization of diagnostics and enhanced sample preparation strategies for rapid detection of foodborne pathogens, b) ecology and control of foodborne pathogens, with a special emphasis on understanding and controlling antimicrobial resistance in the food supple and the environment, including wildlife, and c) development and validation of microbial source tracking approaches.
Bledar Bisha
Phone: (307)766-3140
Email: bbisha@uwyo.edu

Kelly Woodruff works as a laboratory technician/laboratory manager for Dr. Bisha. She grew up in Ventura, California. Kelly then attended the University of Wyoming and received her Bachelor of Science in Animal and Veterinary Sciences with an Animal Biology Concentration. During that time she connected with Dr. Hannah Cunningham-Hollinger where she pursued her Master of Science degree in Animal Science with a focus in Animal Genetics. One of her projects was focused on determining relationships between the cow rumen microbiome during different points of gestation to the developing calf rumen microbiome and meconium. Her second project focused on the impacts of late gestation maternal undernutrition in beef cows on the mature cow rumen microbiome throughout gestation and the developing calf rumen microbiome and meconium. Upon graduation in the Fall of 2020 Kelly was recruited by Dr. Bisha to manage the Wastewater Surveillance project as well as his laboratory.
Kelly Woodruff
Email: klomagno@uwyo.edu

Ryan Knuth is a PhD student working with advisors Dr. Bledar Bisha and Dr. Hannah Cunningham-Hollinger. He grew up near Loganville, Wisconsin on a small commercial Polypay flock and then attended the University of Wisconsin-Platteville to complete a Bachelor of Science in Animal Science. Ryan then completed a Master of Science degree in Animal and Range Sciences under Dr. Tom Murphy at Montana State University. He is now in his doctoral program, where he investigates microbial communities in the ewe, lamb, and environment and any relationships with health, specifically ewe mastitis, and animal performance. Once he completes his program, Ryan hopes to pursue a career in academia to help better the American sheep industry and continue his interests in research, teaching, and outreach. He anticipates graduating in Summer 2022.
Ryan Knuth
Email: rknuth@uwyo.edu
Codi Jo Broten joined the Bisha lab in the Spring of 2017 to begin work on the development and optimization of paper-based microfluidic devices (µPADs) as a platform for the rapid detection of foodborne bacterial pathogens. Before joining the Bisha lab, Codi Jo graduated from Hillsdale College in Hillsdale, Michigan with a Bachelor of Science in biology in May of 2016. As an undergraduate, Codi Jo gained an interest in applied microbiology with her thesis “Evaluation of forskolin for desmutagenic and antioxidant activity via the SOS chromotest and DPPH assay.” Her background at a liberal arts college has helped her to tackle research problems as a graduate student.
As a graduate student, Codi Jo has focused on the detection of Listeria monocytogenes, a foodborne pathogen that disproportionally affects pregnant women and immunocompromised individuals. In the lab, she enjoys the developmental aspect of her work and troubleshooting to find an appropriate solution. Serving as a teaching assistant is also a task that she enjoys, as it provides the opportunity to share the importance of food safety with others.
Codi Jo will graduate with her PhD in 2022. When she is not writing or working in the lab, Codi Jo enjoys hiking and living a healthy lifestyle.
Codi Broten
Email: cbroten@uwyo.edu

Alexys McGuire is currently a master’s student working in Dr. Bisha’s laboratory. She is originally from Akron, CO, but attended the University of Wyoming where she got her bachelor’s degree in Molecular Biology. Right now, she is working on analyzing wastewater for SARS-CoV-2, which is the basis of my master’s research where she is working towards receiving her master’s degree in Animal and Veterinary Science. After she graduates with her master’s degree, she hope to get my PhD and work either in her own independent laboratory or work for the government studying neurodevelopmental disorders. Her expected graduation is Spring 2022.
Alexys McGuire
Email: amcguir9@uwyo.edu

Clara has joined Dr. Bledar Bisha’s lab as a master’s student working to uncover how viral indicators can be best used in monitoring water quality following fecal contamination. Much of her work revolves around the microbial source tracking methodology which makes use of indicators that exhibit host-specificity and enables elucidation of likely sources of contamination. Further, these viral indicators can act as surrogates for assessing pathogen risk and have the potential to inform remediation efforts for impacted waters. She also hopes to further evolve existing methods of molecular quantitation, namely digital PCR, in their application of detecting low-concentration viral species.
Clara grew up in Sheridan, Wyoming. She completed her undergraduate degrees as a double major in Microbiology and Molecular biology at the University of Wyoming in 2021. She was initially drawn to microbiology out of interest for the human microbiome but has since also developed an interest in ecological and pathogenic microbiology. She has also maintained an interest in human medicine and plans to attend medical school following the completion of her graduate degree. Clara’s expected gradation is Spring 2023.
Clara Bouley
Email: cbouley@uwyo.edu
