Chemical Biology
Metabolomics/Lipidomics of bacteria:
This evolutionary biology project aims at elucidating why some bacteria make sterols

Research project funded by the National Science Foundation, NSF-IOS:
"Why do bacteria make sterols?", PI: N. Ward, co-PI: F. Basile
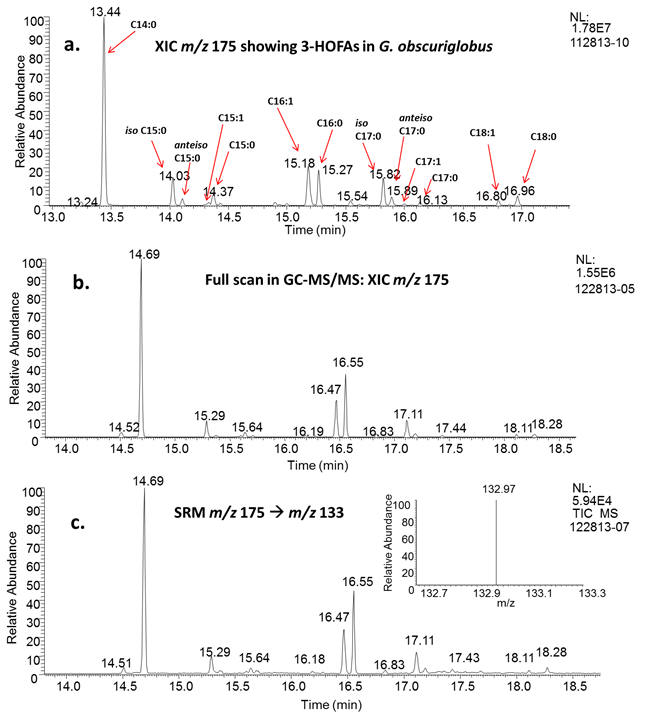
Our laboratory implements several MS techniques (GC-MS, GC-MS/MS, LC-ESI-MS/MS and
LC offline MALDI-MS/MS) for the analysis of lipids, carbohydrates and metabolites
in microorganisms. Recent work from our laboratory identified several hydroxy-fatty
acids (some believed to be associated with lipid A) and polyunsaturated fatty acids
in the microorganism Gemmata obscuriglobus. (Project in collaboration with Dr. Naomi Ward; Read the article in J. Bacteriology, 2016, 198, 221-236 journal link).
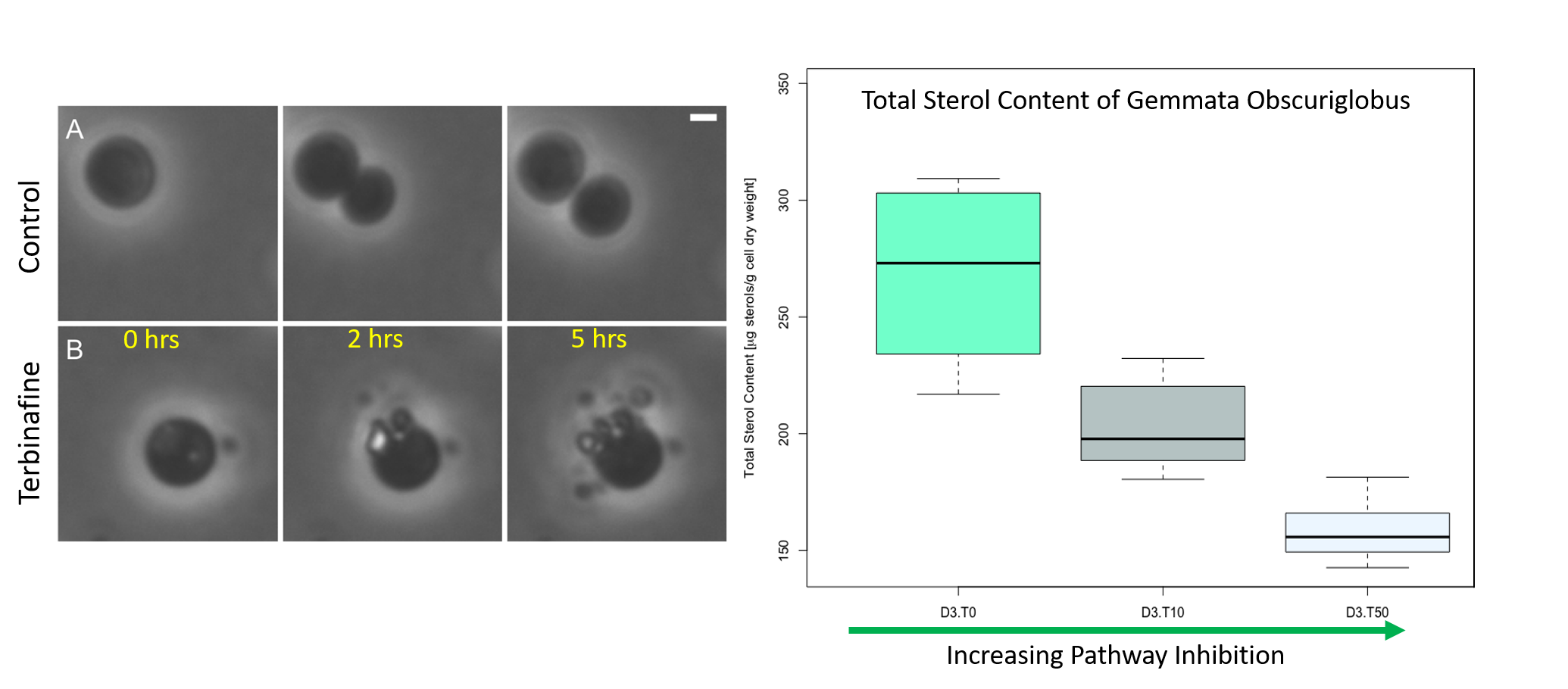
Terbinafine treatment, a suppresor of sterol synthesis, in Gemmata obscuriglobus inhibits normal budding replication (left). Quantitation of sterols in G. obscuriglobus after being treated with terbinafine (right).
Figures from: "Essentiality of sterol synthesis genes in the planctomycete bacterium
Gemmata obscuriglobus", Elena Rivas-Marin, Sean Stettner, Ekaterina Y. Gottshall,
Carlos Santana-Molina, Mitch Helling, Franco Basile, Naomi L. Ward & Damien P. Devos,
Nature Communications | (2019) 10:2916 | https://doi.org/10.1038/s41467-019-10983-7
MICROBESTIARY: Understand science thourgh art Microbestiary hopes to show, in welcoming and memorable ways, that the bustling microbial world
is populated by strange and beautiful characters that are themselves quite charismatic
Microbial Proteomics:
A Study on Strategies for Forming Multicellular Organisms via Cell Aggregation
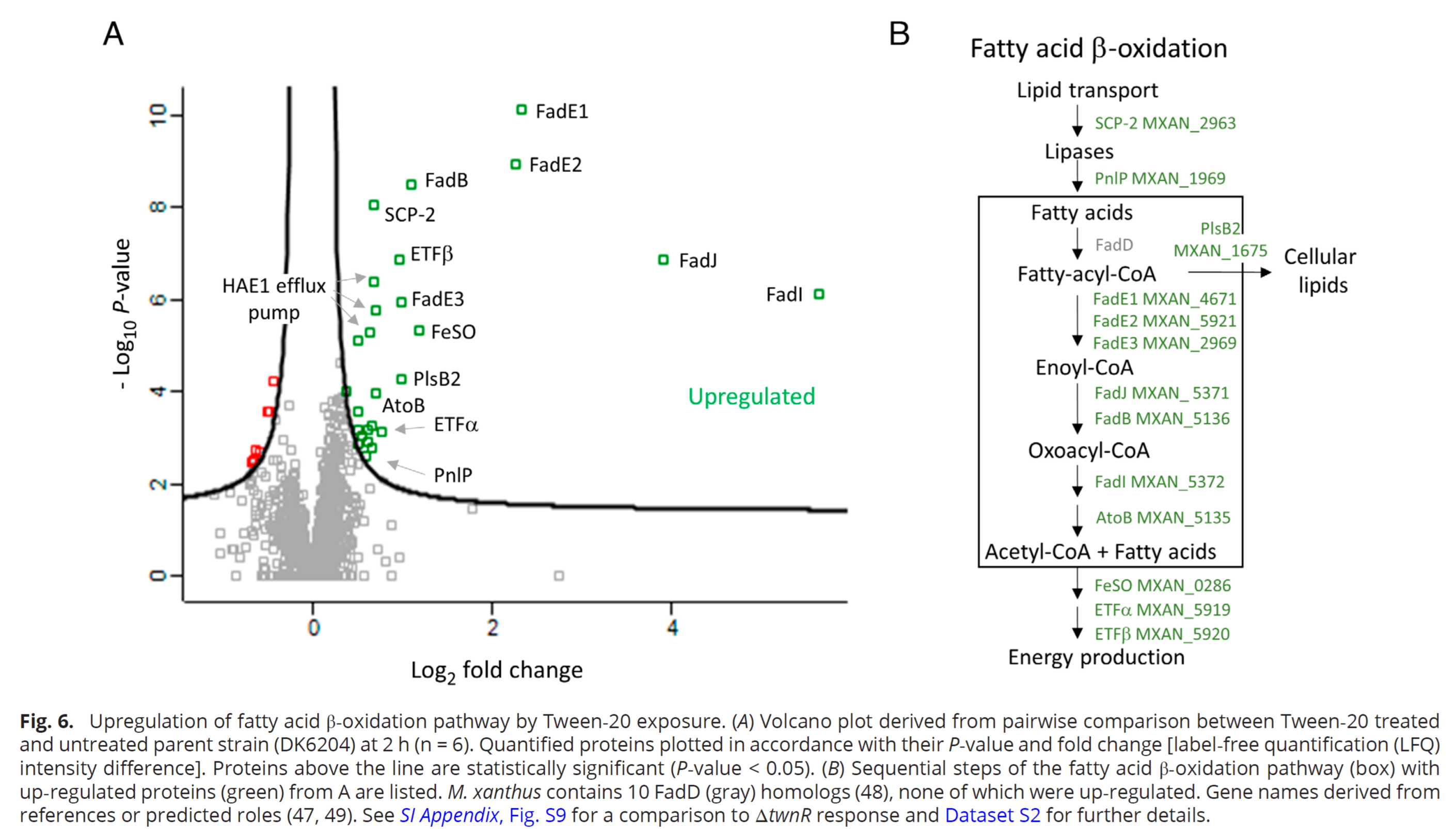
To form multicellular organisms via cell aggregation, cells must distinguish self
from nonself to form a cooperative tissue. In this study, myxobacteria serve as model
organisms for making these transitions. In this project we tested the role of the
TraAB kin recognition system for a role in cell–cell cooperation because these cell
surface receptors mediate the bidirectional exchange of proteins and lipids between
cells. Strikingly, cells that adapted to environmental stresses shared their beneficial
trait with naïve kin in a TraAB-dependent manner. Surprisingly, donor cells also benefited
by apparently establishing harmony in the population when confronted with stress.
We conclude that TraAB plays diverse roles in myxobacterial multicellular behaviors
including their ability to cooperate.
Subedi, Kalpana, Pravas C. Roy, Brandon Saiz, Franco Basile, and Daniel Wall. “Cell–Cell
Transfer of Adaptation Traits Benefits Kin and Actor in a Cooperative Microbe.” Proceedings of the National Academy of Sciences 121, no. 30 (July 23, 2024): e2402559121. https://doi.org/10.1073/pnas.2402559121
This work was supported by grants from the National Institute of General Medical Sciences
R35GM140886 to D.Wall and P20GM103432 from the NIH. Acquisition of the high-resolution
LC-MS/MS instrument was funded by a NSF EPSCoR RII Track-1 grant (NSF 1655726).





