Developing genetic and computational tools for studies in worms
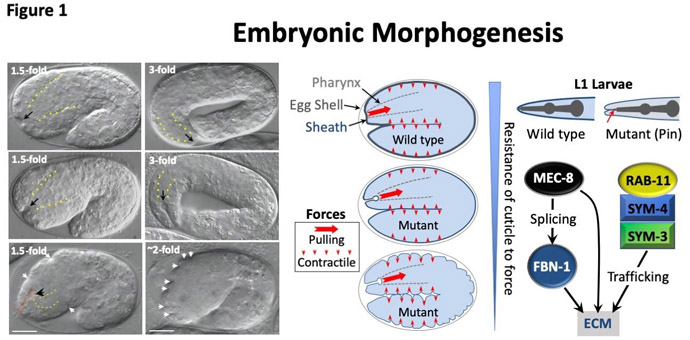
As part of our research program, we are working to develop tools and procedures that may be of use to the greater scientific community. Some of these have involved genetic strategies and bioinformatic pipelines to help expedite the identification of the relevant mutated genes inspecific mutant worms. When scientists carry out genetic screens to obtain mutants, such as those causing defects in molting or embryogenesis, they are generally most interested in determining which one of the 20,000 possible genes contained the mutation that caused the observed defect. This is a bit of a needle in a haystack problem and can be compounded by a number of complicating factors. One of our innovations was to develop a strategy termed the Sibling Subtraction method, which can help to streamline this process (Figure 2).
More recently, we developed a computational tool to help scientists with their CRISPR experiments. CRISPR is a tool used to make edits to the genomes of cells and animals, and its discovery and development led to the 2020 Nobel prize in chemistry. Our tool, called CRISPR cruncher, aids scientists in the design of CRISPR experiments so that they can identify useful genomic edits more efficiently. This tool was developed in collaboration with the Wyoming INBRE Bioinformatics Core, which was also instrumental in our development of the Sibling Subtraction method.