Contact Us




Dr. Mark Gomelsky
Agriculture Bldg, Room 6007
Department of Molecular Biology
University of Wyoming
Department 33944
1000 E. University Ave.
Laramie, WY 82071
Phone: (307) 766-3522
Email: gomelsky@uwyo.edu
Research Interests
The Gomelsky Lab has several foci of interest, including Optogenetics in near-infrared, Listeria-plant biofilms and Listerial bactodrones as cancer therapy.
Optogenetics in near-infrared
Light within the near-infrared optical window (~670-900 nm) penetrates mammalian tissues much deeper than visible light; therefore, it can activate engineered cells located under the skin or skull (Fig. C). To "teach" mammalian cells how to respond to the near-infrared light, we are employing bacteriophytochromes, a class of natural photoreceptors (Fig. A&B). We have deciphered principles of engineering homodimeric bacteriophytochromes to guide construction of protein chimeras, comprising the bacteriophytochrome photosensory module and the desired output module. One example is an engineered chimeric near-infrared light-activated adenylate cyclase (Fig. B). It can increase insulin production in mammalian cells or suppress cAMP-dependent neuronal waves in mice (Fig. B&C). We intend to optimize the near-infrared light-activated adenylate cyclase for optogenetic applications in diabetes type I and in neuronal regulation. We are also engineering new homodimeric bacteriophytochrome enzymes to control activities of mammalian cells noninvasively, via externally localized sources of near-infrared light.
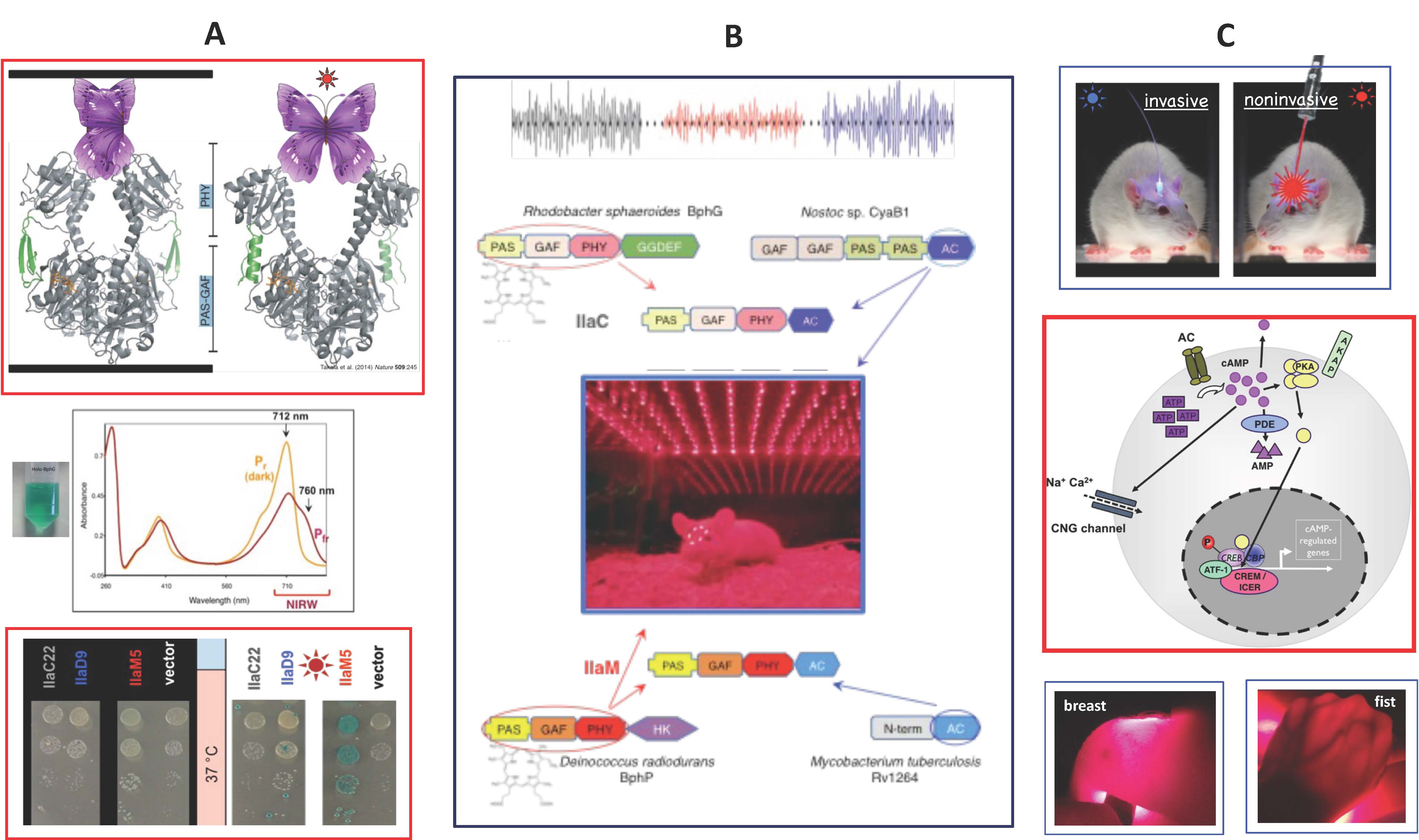
 To learn more, see:
To learn more, see:
Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen
DM, Gomelsky M. 2014. Engineering adenylate cyclases regulated by near-infrared window light. Proc
Natl Acad Sci USA 111:10167-72.
https://www.pnas.org/doi/abs/10.1073/pnas.1324301111
Ryu MH, Gomelsky M. 2014. Synthetic second messenger module controlled by near-infrared window light.
ACS Synth Biol 3:802-810.
https://pubs.acs.org/doi/abs/10.1021/sb400182x
Fomicheva A, Zhou C, Sun QQ, Gomelsky M. 2019. Engineering adenylate cyclase activated by near-infrared window light for mammalian
optogenetic applications. ACS Synth Biol 8:1314-1324.
https://pubs.acs.org/doi/abs/10.1021/acssynbio.8b00528
Listeria-plant biofilms
Fresh produce contaminated with the foodborne pathogen, Listeria monocytogenes, has caused major listeriosis outbreaks in the USA in the past decades. The 2011 cantaloupe-associated outbreak was the highest mortality food-poisoning incident in the USA history in almost a century. Despite the growing prevalence of fruits and vegetables as sources of listerial contamination, we know little about how Listeria colonizes them, which hinders our ability to prevent fresh produce contamination with Listeria.
We discovered that Listeria synthesizes a novel exopolysaccharide (EPS), {4)-β-ManpNAc-(1-4)-[α-Galp-(1-6)]-β-ManpNAc-(1-}, where ManpNAc is N-acetylmannosamine and Galp is galactose, and hypothesized that it is involved in listerial growth and survival on fresh produce. The pss operon contains the genes for EPS biosynthesis. The Pss synthetic machinery is activated by the second messenger, c-di-GMP (Fig. A). Figs B&C show that the EPS is synthesized by Listeria on plant surfaces and increases colonization. In this project we want to understand regulation of Listeria biofilm formation on plant surfaces, to characterize the role of the EPS-biofilms in listerial colonization and long-term survival on fresh produce, and to devise the means of preventing biofilm formation. For example, one natural, plant-derived compound blocks listerial colonization (Fig. C).
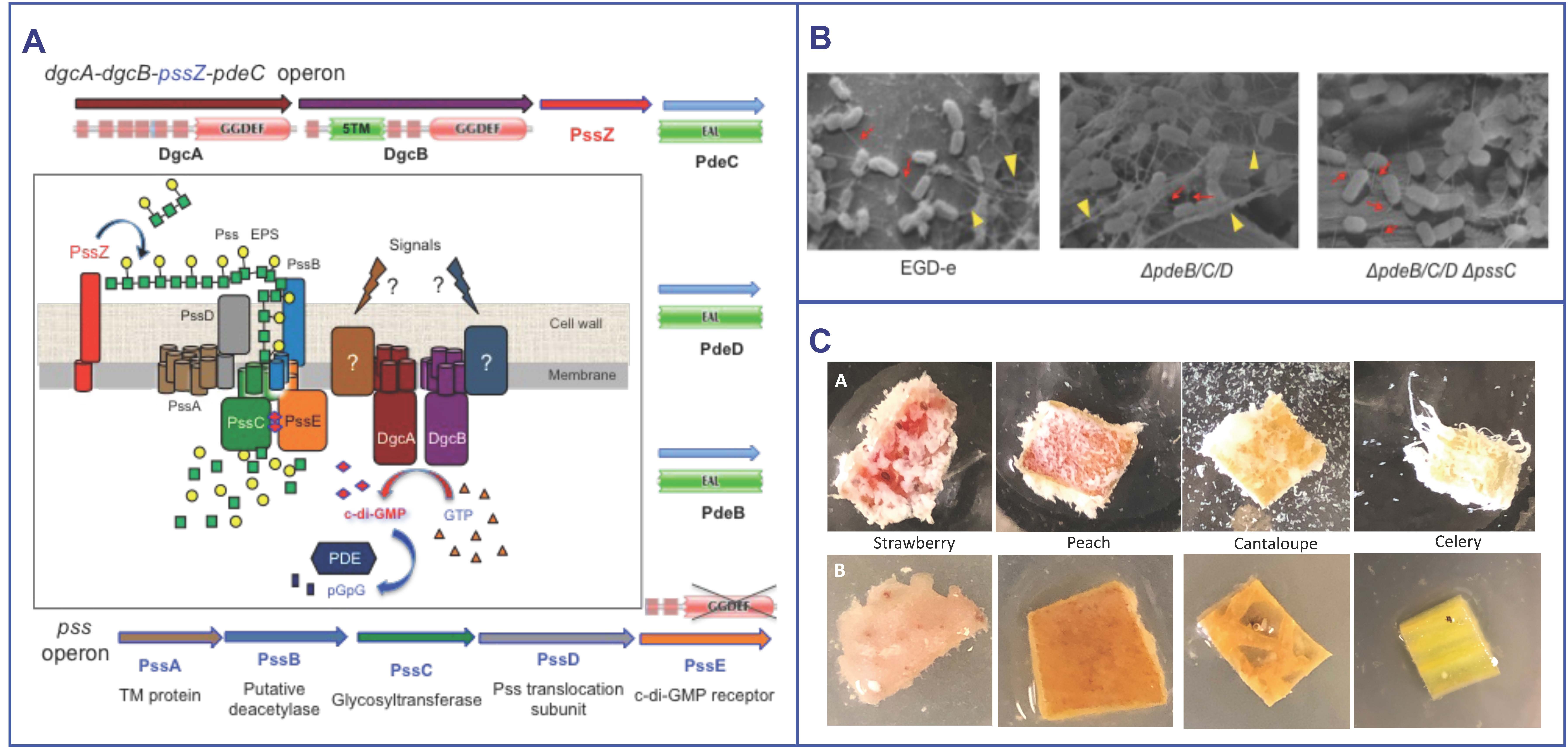
 To learn more, see:
To learn more, see:
Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger.
Microbiol Mol Biol Rev 77: 1-52.
https://journals.asm.org/doi/full/10.1128/MMBR.00043-12
Chen LH, Köseoğlu VK, Güvener ZT, Myers-Morales T, Reed JM, D’Orazio SEF, Miller KW,
Gomelsky M. 2014. Cyclic di-GMP-dependent signaling pathways in the pathogenic firmicute Listeria
monocytogenes. PLoSPathog 10:e1004301.
http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1004301
Köseoğlu VK, Heiss C, Azadi P, Topchiy E, Güvener ZT, Lehmann TE, Miller KW, Gomelsky
M. 2015. Listeria monocytogenes exopolysaccharide: origin, composition, biosynthetic
machinery, and c-di-GMP-dependent regulation. Mol Microbiol 96:728-43.
https://onlinelibrary.wiley.com/doi/10.1111/mmi.12966
Listerial bactodrones as cancer therapy
The attenuated strains of Listeria monocytogenes injected intraperitonially are phagocytosed by myeloid-derived suppressor cells (MDSCs), which deliver them to primary tumors and metastases (Fig. A, B). In the immune suppressive tumor microenvironment, Listeria propagates and spreads into tumor cells. We are employing the tools of synthetic microbiology to turn Listeria into remotely controlled bacterial drones, bactodrones, that can deliver various therapeutic payloads to tumor and immune cells and release them upon activation (Figs. A, C, D). Our goal is to engineer a fleet of listerial bactodrones to effectively target various cancers.
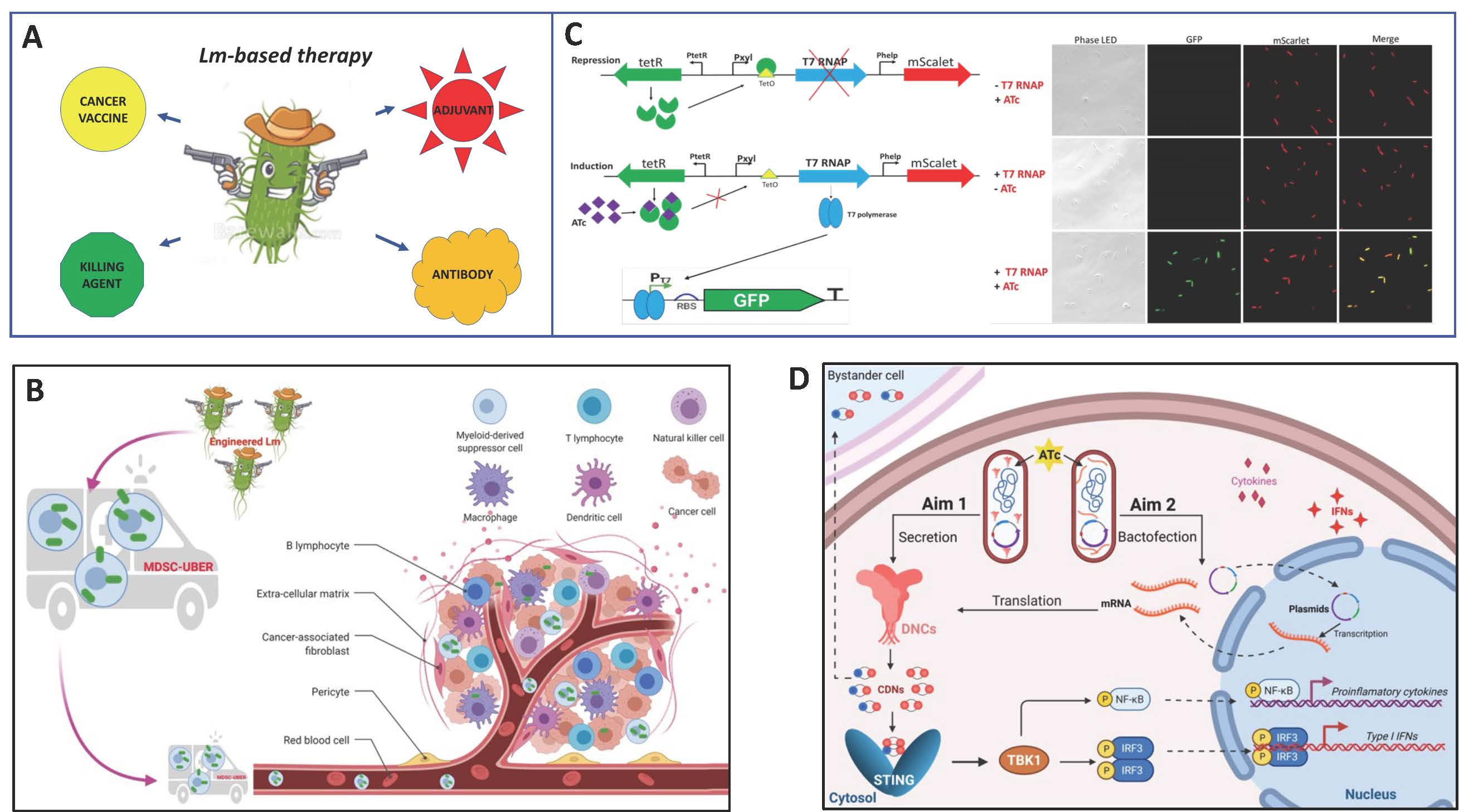
Metabolic and cellular engineering
Most bacteria are believed to divide generating two identical daughter cells. However, a simple polar-localized signaling system (red dot) can convert bacterial cell division into asymmetric division. The ability to generate daughter cells that have different fates and functions (grey with a red pole and green without a pole) is of significant interest in microbial biotechnology, because it offers a way to separate cell growth and product synthesis.
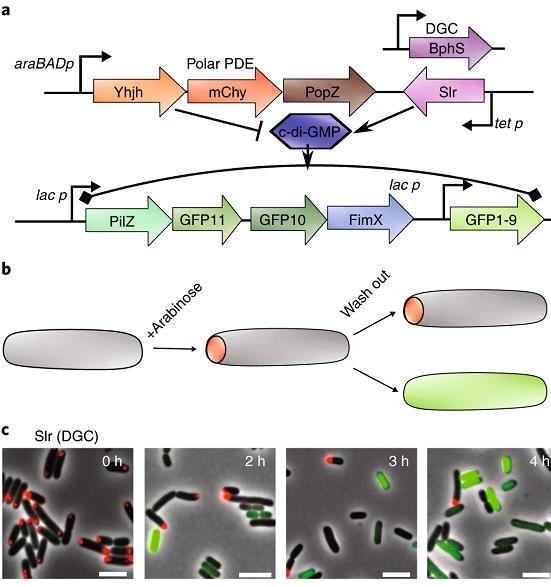
To learn more, see:
Mushnikov NV, Fomicheva A, Gomelsky M, Bowman G. 2019. Inducible asymmetric cell division and cell differentiation in a bacterium.
Nat Chem Biol 15:925-931.
https://www.nature.com/articles/s41589-019-0340-4
Cyclic dimeric GMP, c-di-GMP, is a broadly distributed second messenger that controls biofilm formation, motility and virulence in diverse bacteria. We engineered optogenetic systems to manipulate bacterial behavior by increasing or lowering c-di-GMP levels with red and blue light.

To learn more, see:
Ryu MH, Fomicheva A, Moskvin OV, Gomelsky M. 2017. Optogenetic module for dichromatic control of c-di-GMP signaling. J Bacteriol
199:e00014-17.
https://journals.asm.org/doi/10.1128/JB.00014-17
O’Neal L, Ryu MH, Gomelsky M, Alexandre G. 2017. Optogenetic manipulation of c-di-GMP levels reveals the role of c-di-GMP in
regulating aerotaxis receptor activity in Azospirillum brasilense. J Bacteriol 199:e00020-17.
https://journals.asm.org/doi/10.1128/JB.00020-17
Ryu MH, Fomicheva A, O’Neal L, Alexandre G, Gomelsky M. 2017. Using light-activated enzymes for modulating intracellular c‑di-GMP levels
in bacteria. Methods Mol Biol 1657:169-186.
https://link.springer.com/protocol/10.1007/978-1-4939-7240-1_14
Hydrogen is a clean fuel. Anoxygenic phototrophic bacteria can utilize solar energy and organic waste products to synthesize large quantities of hydrogen. We used metabolic maps, flux balance analysis and global gene regulatory systems of a model bacterium Rhodobacter sphaeroides to engineer a high hydrogen producing strain.
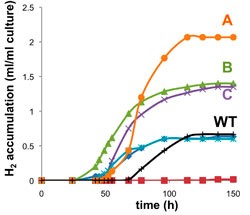
To learn more, see:
Ryu MH, Hull NC, Gomelsky M. 2014. Metabolic engineering of Rhodobacter sphaeroides for improved hydrogen production.
Intl J Hydrogen Energy 39:6384-6390.
http://www.sciencedirect.com/science/article/pii/S0360319914003814
Gomelsky M, Zeilstra-Ryalls JH. 2013. The living genome of a purple nonsulfur photosynthetic bacterium: Overview
of the Rhodobacter sphaeroides transcriptome landscapes. Adv Botanical Res 66:179-203.
http://www.sciencedirect.com/science/article/pii/B9780123979230000060
Moskvin OV, Bolotin D, Wang A, Ivanov PS, Gomelsky M. 2011. Rhodobase, a meta-analytical tool for reconstructing gene regulatory networks
in a model photosynthetic bacterium. BioSystems 103: 125-131.
http://www.sciencedirect.com/science/article/pii/S0303264710002030
Contact Us




Dr. Mark Gomelsky
Agriculture Bldg, Room 6007
Department of Molecular Biology
University of Wyoming
Department 33944
1000 E. University Ave.
Laramie, WY 82071
Phone: (307) 766-3522
Email: gomelsky@uwyo.edu
