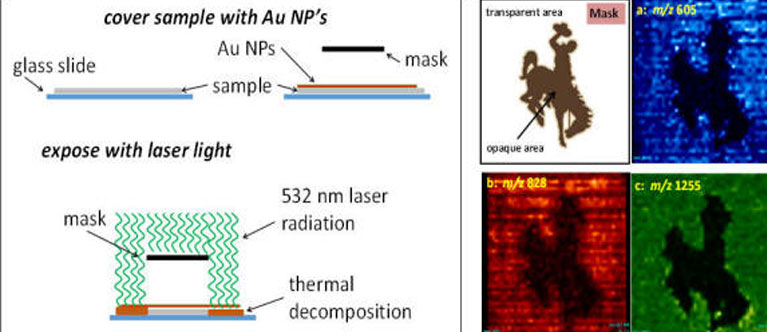
"the most difficult portion of the analytical process is that involving isolation
of the desired constituent in a measurable state. The actual process of measurement
is simple."
from: Ernest B. Sandell and Phillip J. Elving, Methods of Analytical Chemistry, Chapter
1 (p. 7) in Treatise on Analytical Chemistry Part I Theory and Practice; I. M. Kolthoff
and Phillip J. Elving (Eds.), 1959
Our Research
Funding for this research has been provided by:
|
|
|
|
Contact Information
Franco Basile, Ph.D.
Dept 3838
Department of Chemistry
1000 E. University Ave.
University of Wyoming
Laramie, WY 82071
Office (307) 766-4376
Laboratory (307) 766-2991
basile@uwyo.edu
- Basile Faculty Page
- Department of Chemistry
- Our laboratory is located in the Physical Sciences Bldg, Room 444 (Online Map|Printable Map)
- UW campus parking permit map