Daniel Wall, Ph.D.
Department of Molecular Biology
Professor and Chair

Research Overview
A fundamental question in biology is how individual cells within a multicellular organism
interact to coordinate diverse processes? We are interested in this question from
molecular and evolutionary perspectives.
To address this topic our laboratory studies the genetically tractable organism called Myxococcus xanthus. As a group, myxobacteria are highly social microbes that move, hunt and develop as coherent multicellular units. Their most notable behavior is their ability to build fruiting bodies in response to starvation.
Here, thousands of individual and small groups of cells coalesce from their environment to assemble fruiting bodies wherein vegetative cells differentiate into different cell types including environmentally resistant spores (Fig. 1). In this regard our laboratory is focused on two general questions: i) At a molecular level how do cells recognition their clonemates and discriminate against non-kin? ii) How, in mechanistic and evolutionary terms, can myxobacteria transition between solitary and multicellular lifestyles?
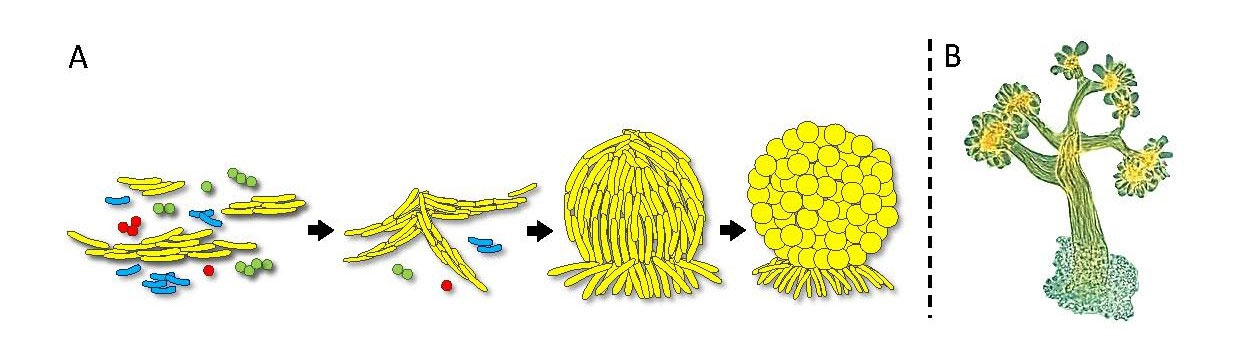
Fig. 1. Fruiting body development. A) A diagram of cell aggregation and fruiting body formation by M. xanthus cells (yellow) from a diverse microbial community. B) Micrograph of an actual Chondromyces crocatus fruiting body.
We propose that a partial answer to the above questions lies within a novel process that we discovered and named – outer membrane exchange (OME). Here, upon physical contact with a neighboring cell the outer membranes transiently fuse and cells exchange outer membrane proteins and lipids (Fig. 2). ME is mediated by a duo of cell surface proteins, TraA and TraB, which act as cell-cell adhesins and fusagens. The exchange or sharing of significant amounts of cellular resources within diverse microbial communities (soil) raises questions about how partner cells are identified and whether these interactions are regulated. Indeed, we found that OME is highly selective and only occurs between cells that contain identical or nearly identical TraA proteins (Fig. 3). Therefore OME involves kin recognition that is governed by genetically defined homotypic interactions between polymorphic traA alleles. Thus cells that express identical or nearly identical traA alleles form a functional recognition group, while strains with divergent TraA receptors will not exchange (Fig. 4). Molecular and phylogenetic studies further suggest that among environmental populations there are hundreds of different TraA-derived recognition or social groups (Fig. 3B).
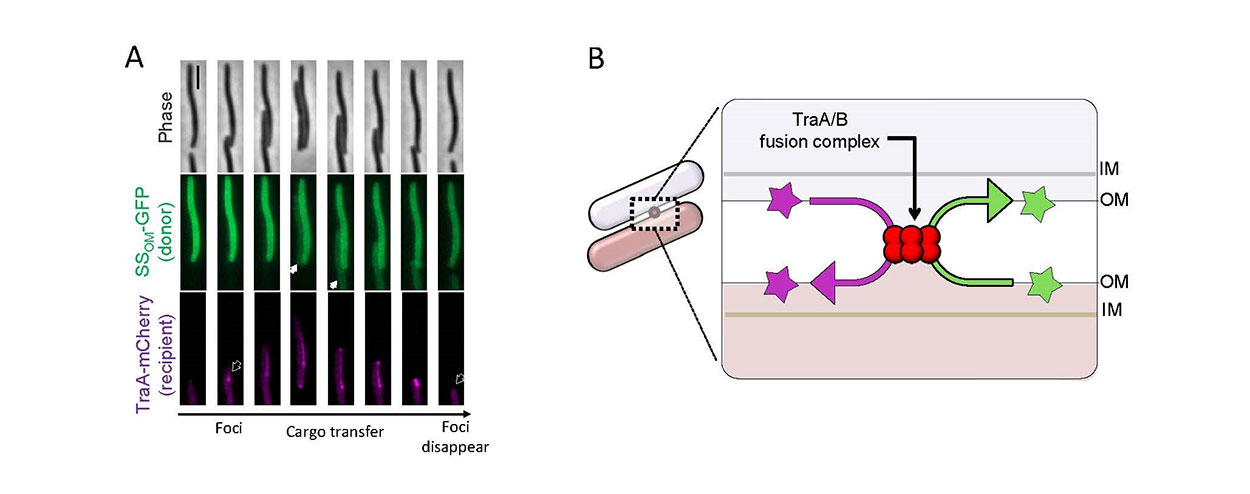
Fig. 2. Outer membrane exchange (OME) in myxobacteria. A) Time-lapse series of two cells making contact by gliding motility followed by TraA-TraA recognition (foci formation, arrow outline left) and then cargo transfer (GFP with lipoprotein signal sequence, white arrow) from the ‘donor’ to ‘recipient’ cell. The recipient cell expresses a TraA-mCherry fusion reporter that coalesces upon cell-cell contact. Note that the recipient cell reverses direction and once the cells disengage foci disappear (arrow outline right). Scale bar 1 μm. B) Schematic of OME where two adjacent cells recognizing each other by homotypic TraA interactions to form a TraAB OM fusion junction. Subsequently, transient membrane fusion allows the passive diffusion of proteins and lipids bidirectionally between cells. OM, outer membrane; IM, inner membrane.
Phenotypically, Tra-dependent OME leads to both beneficial and adversarial outcomes.
For instance, under certain conditions damaged proteins or lipopolysaccharides in
the cell envelop can be replaced and repaired by healthy partner cells, representing
a helping cooperative behavior. In contrast, under different circumstances OME leads
to policing or discrimination behaviors. These latter outcomes are driven by the exchange
of a suite of polymorphic toxins that vary widely between myxobacteria isolates. Here,
clonemate cells will express a suite of immunity proteins, which are not transferred,
and thus confer self-resistance, while non-self-cells that happen to bear compatible
TraA receptors will be poisoned. In summary, OME represents a new paradigm for how
bacterial cells communicate, interact and transition from individuals into functional
multicellular units. Our future studies are aimed at understanding the molecular details
of cell-cell recognition, mechanism of outer membrane fusion and the social and evolutionary
consequences of OME.
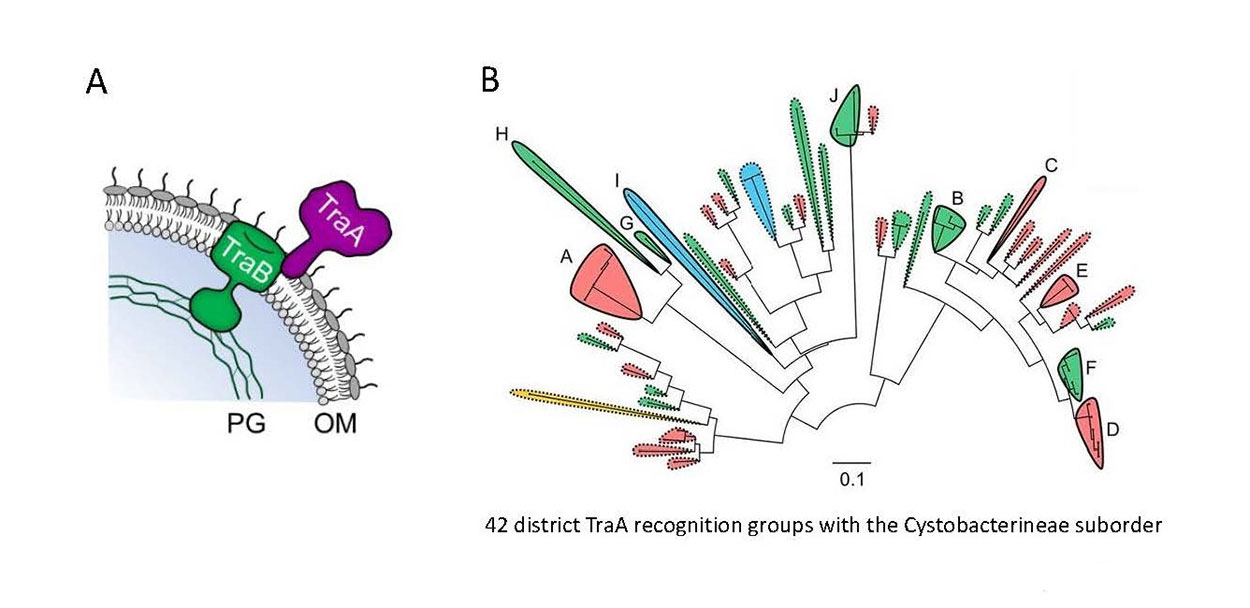
Fig. 3. Self-recognition among a wide range of myxobacteria is governed by sequence variability in the TraA cell surface receptor. A) Model of the TraAB proteins in the cell envelop. B) Maximum likelihood tree consists of 59 myxobacterial TraA orthologs (variable domain). Solid lines indicate experimentally determined recognition groups (letters indicate groups), and dashed lines show predicted recognition groups. Shaded colors indicate key specificity residue at position 205. Scale bar indicates the number of substitutions per amino acid residue.
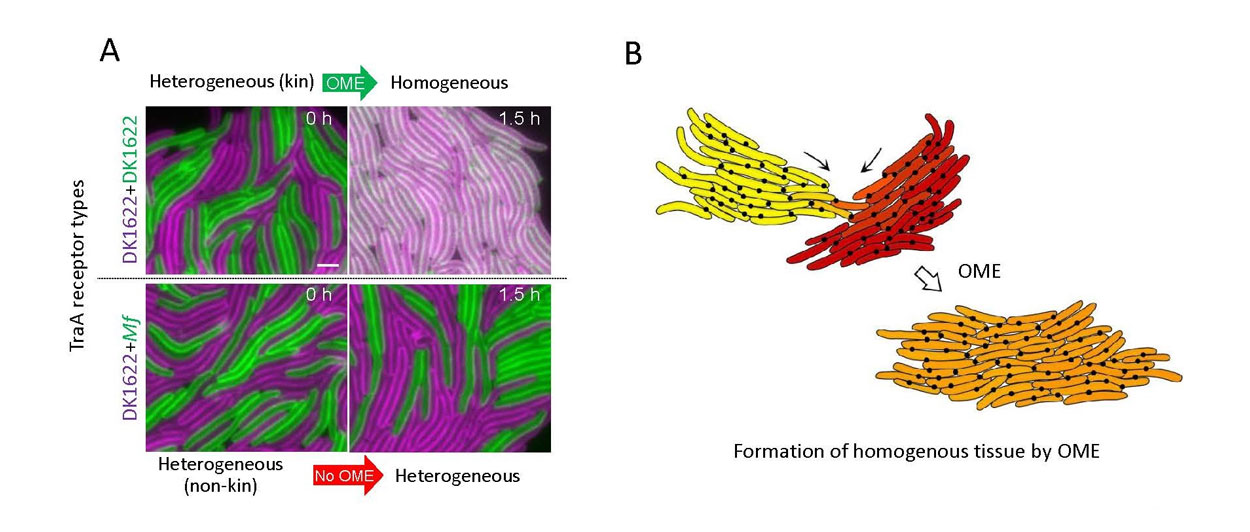
Fig. 4. Tissue formation mediated by OME. A) Isogenic M. xanthus strains were mixed that harbor different fluorescent reporters (SSOM-GFP or SSOM-mCherry cellular cargo), which represent distinct adaptation states caused by growth in different environments. Top panels: Compatible TraA receptors allow exchange after a brief incubation. Bottom panels: In contrast, incompatible receptors prevent exchange. traA alleles shown at the left. Scale bar = 1 µm. B) Illustration of two kin populations that adapted to different microenvironments and are thus phenotypically divergent (top, labeled yellow and red). Following TraA-TraA recognition the groups merge into a homogeneous population or tissue through OME (orange). Black dots represent TraAB foci located at cell–cell contacts that are analogous to gap junctions in eukaryotic tissues.
Education
- Postdoctoral Fellow, Stanford University, 1998
- Ph.D., University of Utah, 1994
- B.A., Sonoma State University, 1988
Professional Experience
- Principal Scientist, Anadys Pharmaceuticals (acquired by Roche), 2006
- Senior Scientist, Elitra Pharmaceuticals (acquired by Merck), 2002
Recent Publications
- Subedi, K., Roy, P., Saiz, B., Basile, F. and Wall, D. 2024. Cell-cell transfer of adaptation traits benefits kin and actor in a cooperative microbe. PNAS 121 (30): e2402559121.
- Kaimer, C., Weltzer, M.L. and Wall, D. 2023. Two reasons to kill: Predation and kin discrimination in myxobacteria. Invited/peer review, Microbiology, 10.1099/mic.0.001372
- Weltzer, M. and Wall, D. 2023. Social diversification driven by mobile genetic elements. Genes 14(3), 648: 10.3390/genes14030648
- Zhang, L., Dong, C., Wang, J., Liu, M., Wang. J., Hu J., Liu, L., Liu, X., Xia, C., Zhong, L., Zhao, Y., Ye, X., Huang, Y., Fan, J., Cao, H., Wang, J., Li, Y., Wall, D., Li, Z., Cui, Z. 2023. Predation of oomycetes by myxobacteria via a specialized CAZyme system arising from adaptive evolution. ISME J. 17:1089-1103
- Subedi, K. and Wall, D. 2022. Conditional and synthetic type IV pili-dependent motility phenotypes in Myxococcus xanthus. Front Microbiol. fmicb.2022.879090.
- Vassallo, C., Sah, G., Weltzer, M. and Wall, D. 2021. Modular outer membrane exchange toxins exploit discrete cell entry pathways. mBio, 12:e02388-21.
- Balagam, R., Cao, P., Sah, G.P., Zhang, Z., Subedi, K., Wall, D. and Igoshin, O.A. 2021. Emergent myxobacterial behaviors arise from reversal suppression induced by kin contacts. mSystems,6:6 e00720-21.
- Vassallo, C., Troselj, V., Weltzer, M. and Wall, D. 2020. Rapid diversification of wild social groups driven by toxin-immunity loci on mobile genetic elements. ISME J 14:2474-2487.
- Sah, G. and Wall, D. 2020. Kin recognition and outer membrane exchange (OME) in myxobacteria. Current Opinion Microbiology 56:81-88.
- Sah, G., Cao, P. and Wall, D. 2020. MYXO-CTERM sorting tag directs proteins to the cell surface via the type II secretion system. Molecular Microbiology 113:1038-1051.
- Cao, P. and Wall, D. 2020. The fluidity of bacterial outer membrane is species specific: Bacterial lifestyles and the emergence of a fluid outer membrane. BioEssays 42, 1900246.
- Troselj, V., Pathak, D. and Wall, D. 2020. Conditional requirement of SglT for type IV pili function and S-motility in Myxococcus xanthus, Microbiology, 166:349-358.
- Cao, P. and Wall, D. 2019. Direct visualization of a molecular handshake that governs kin recognition and tissue formation in myxobacteria. Nature Communications 10, 3073.
- Cao, P, Awal, R.P., Müller, R. and Wall, D. 2019. A highly polymorphic receptor governs many distinct self-recognition types within the Myxococcales order. mBio 10:e02751-18.
- Vassallo, C. and Wall, D. 2019. Self-identity barcodes encoded by six expansive polymorphic toxin families discriminate kin in myxobacteria. PNAS 116 (49): 24808-24818.
- Troselj, V., Treuner-Lange, A., Søgaard-Andersen, L. and Wall, D. 2018. Physiological heterogeneity triggers sibling conflict mediated by type VI secretion system in an aggregative multicellular bacterium. mBio 9:e01645-17.
- Kitamura, S., Owensby, A. Wall, D. and Wolan, D. W. 2018. Novel lipoprotein signal peptidase inhibitors with antibiotic properties identified through design of a robust in vitro HT platform. Cell Chem. Biol. 25:1-8.
- Troselj, V., Cao, P and Wall, D. 2018. Cell-cell recognition and social networking in bacteria. Environ. Microbiol. 20(3), 923–933.

